1. Introduction
Cyanobacteria and microalgae occupy a unique and indispensable position in Earth’s biosphere. As primary producers, these photosynthetic microorganisms are responsible for nearly half of global oxygen production and carbon dioxide fixation, forming the foundation of both aquatic and terrestrial food webs (Burgunter-Delamare et al., 2024; Mazard et al., 2016). Historically, their study was constrained by classical taxonomic approaches, relying on morphological characteristics observed under light microscopy. While these methods allowed initial classification, they overlooked the vast hidden diversity that could not be cultivated in laboratory conditions, including species embedded in dense exopolymeric sheaths or engaged in obligate symbiotic relationships (Zammit et al., 2023). As such, the term “microbial dark matter” has emerged to describe the largely uncharacterized reservoir of microbial diversity, emphasizing the limitations of traditional cultivation-dependent techniques.
The past decade, however, has witnessed a profound transformation in the study of these organisms. Advances in systems biology, high-resolution imaging, and metabolomics have enabled researchers to examine cyanobacteria and microalgae beyond their morphology, revealing their intricate physiological, ecological, and biochemical properties. These tools have unlocked new opportunities to explore biodiversity in previously inaccessible and extreme environments, including polar ice sheets, deep-sea hydrocarbon seeps, and mesophotic reefs (Gusmão et al., 2023; Lauritano et al., 2020; Zammit et al., 2023). Remote sampling via robotics and remotely operated vehicles (ROVs) allows scientists to collect and maintain samples at depths up to 6000 meters, preserving the in situ physiological state essential for accurate characterization (Garel et al., 2019). Such technological breakthroughs have shifted the paradigm from observational ecology to mechanistic understanding, enabling scientists to bridge the gap between laboratory models and natural ecosystems.
The urgent global challenges posed by climate change have further intensified research in this field. Rising sea surface temperatures, ocean acidification, and nutrient loading have contributed to the increased frequency and toxicity of harmful algal blooms (HABs), events that can devastate marine biodiversity, fisheries, and public health (Tsui & Kong, 2023; IPCC, 2019). In response, researchers have embraced high-resolution environmental monitoring platforms to track these dynamic ecosystems in real time. Autonomous Surface Vehicles (ASVs) equipped with multi-sensor sondes and bathymetric sonar facilitate continuous observation of water quality parameters, including chlorophyll-a and phycocyanin concentrations, allowing early detection of bloom events (Salman et al., 2022). Additionally, the Environmental Sample Processor (ESP) performs in situ molecular analyses of toxin-producing species such as Pseudo-nitzschia, providing rapid insights into HAB composition and toxicity (Moore et al., 2021). These approaches exemplify the integration of robotics, high-throughput sampling, and molecular biology to address urgent ecological and public health concerns.
Robotic systems have revolutionized the discovery and monitoring of cyanobacteria and microalgae in extreme habitats. ROVs fitted with precision sampling arms and acrylic core samplers have enabled direct access to deep-sea asphalt ecosystems, revealing cyanobacterial taxa previously thought to inhabit only sunlit environments (Gusmão et al., 2023; Zammit et al., 2023). In parallel, high-definition imaging combined with laser guidance has documented the proliferation of filamentous biomats, composed of genera such as Lyngbya and Pseudanabaena, which envelop mesophotic reefs and alter local biodiversity patterns (Sellanes et al., 2021). These technologies not only expand our understanding of microbial diversity but also provide insights into ecological interactions and habitat engineering by microbial mats in low-light, high-pressure environments.
Maintaining the physiological integrity of samples is critical for accurate characterization. Pressure-retaining samplers capable of withstanding up to 60 MPa allow for the recovery of deep-sea microorganisms without inducing decompression stress, preserving cellular activity and gene expression profiles for downstream analyses (Garel et al., 2019). In high-throughput laboratory contexts, 3D-printed polycarbonate inoculation stamps facilitate rapid and precise transfer of microbial strains into multi-well plates, enabling systematic studies of symbiotic, antagonistic, and metabolic interactions across diverse taxa (Temkin et al., 2019). Furthermore, in controlled experiments simulating ocean acidification, dialysis bags with molecular weight cut-offs prevent mechanical damage to sensitive species such as Alexandrium, allowing researchers to study physiological responses without confounding turbulence from CO2 bubbling (Tsui & Kong, 2023).
High-speed sampling techniques are essential for accurate metabolomic profiling, as phototrophic microorganisms respond rapidly to environmental shifts. Fast vacuum filtration allows biomass separation from culture media in as little as 10 seconds, effectively quenching metabolism and preserving the in situ physiological state (Obata et al., 2013a; Schwarz et al., 2013). This approach is particularly valuable in studies examining carbon and nitrogen allocation, secondary metabolite production, and stress-response pathways in diverse microalgal species. The integration of metabolomics with proteomics and transcriptomics further elucidates the complex metabolic networks underpinning resilience and adaptation, revealing high-value compounds such as mannitol, storage lipids, and bioactive secondary metabolites with pharmaceutical and industrial applications (Inwongwan et al., 2025; Obata et al., 2013a; Schwarz et al., 2013).
Microscopy has similarly undergone a paradigm shift. Atomic Force Microscopy (AFM) enables the visualization of cellular ultrastructure in near-native conditions, avoiding artifacts induced by fixation or staining. For instance, AFM studies of Dunaliella tertiolecta demonstrate remarkable stability in cell surface morphology across temperature gradients, reflecting adaptive strategies for survival under thermal stress (Novosel et al., 2022). AFM-based variants, including Fluidic Force Microscopy (FluidFM) and AFM-IR, allow precise characterization of cell hydrophobicity and subcellular chemistry, such as lipid droplet formation, providing a direct link between physiological state and metabolic potential Cryo-Electron Microscopy (Cryo-EM) extends this capability, reconstructing three-dimensional architectures of photosystems and phycobilisomes in cyanobacteria and revealing energy transfer mechanisms at near-atomic resolution (Lian et al., 2022; Tatters et al., 2013). Complementary techniques such as Microcrystal Electron Diffraction (MicroED) have illuminated cryptic algaecides produced in microbial symbioses, enhancing our understanding of chemical signaling within the phycosphere (Sun et al., 2011).
These methodological advancements not only support biodiversity discovery but also enable the development of cyanobacteria and microalgae as sustainable biofactories. Systems-level insights into interspecies interactions, metabolite exchange, and co-cultivation dynamics have informed strategies for optimizing the production of biofuels, pigments, and high-value metabolites (Chen et al., 2025; Rojas-Villalta et al., 2023; Saini et al., 2024). By deciphering the chemical language of the phycosphere, researchers can manipulate microbial consortia to enhance growth and metabolic output, promoting industrial scalability while maintaining ecological sustainability. Furthermore, studies of microbial inoculants for waste biomass conversion highlight the potential of cyanobacteria and microalgae in circular bioeconomy frameworks, underscoring their dual role in environmental remediation and resource generation (Kiruba & Saeid, 2022; Yu et al., 2013).
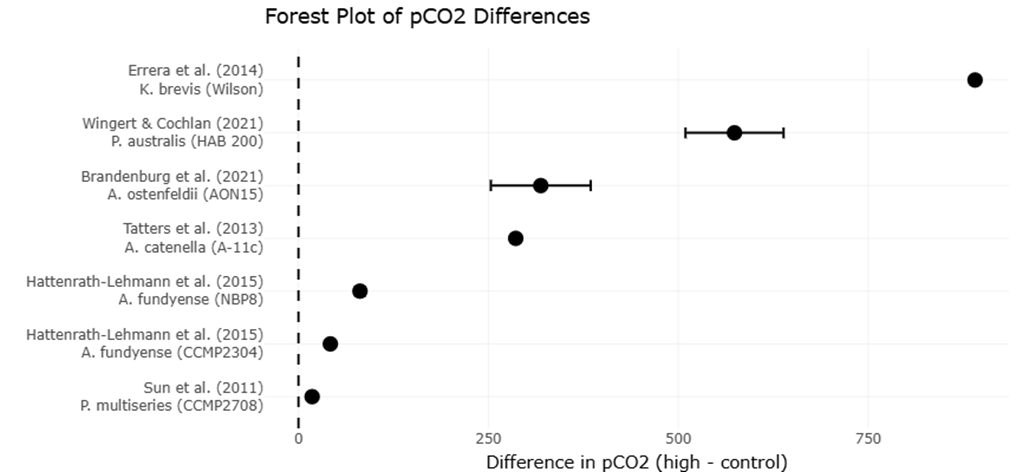
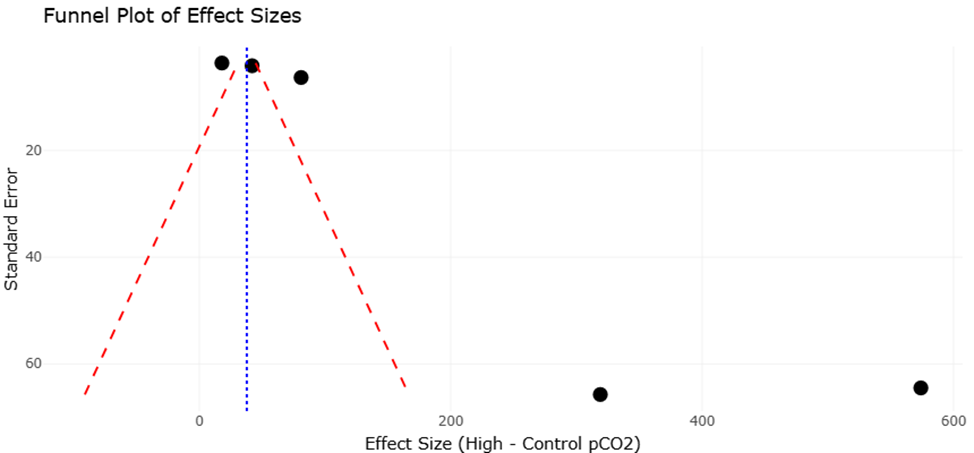
Meta-analyses of physiological responses to elevated pCO2 underscore the nuanced effects of ocean acidification on microalgal growth and toxicity. Across multiple studies, species such as Alexandrium fundyense, A. catenella, and Prorocentrum multiseries demonstrated enhanced growth under elevated CO2, whereas Pseudo-nitzschia australis showed growth inhibition at high pCO2 (Hattenrath-Lehmann et al., 2015; Tatters et al., 2013; Wingert & Cochlan, 2021). Toxin production followed similarly variable trends: several Alexandrium strains exhibited increased saxitoxin synthesis under acidified conditions, while other species like A. ostenfeldii and A. tamarense showed decreased toxicity (Brandenburg et al., 2021; Van de Waal et al., 2014). These results highlight the importance of species-specific assessments when predicting ecological impacts of climate change on harmful algal blooms, emphasizing the value of systematic review and meta-analytic approaches for synthesizing complex datasets.
Collectively, the integration of robotics, advanced sampling, metabolomics, and high-resolution microscopy has transformed our ability to study cyanobacteria and microalgae. By providing a bridge between natural habitats and laboratory analysis, these technologies enable a holistic understanding of microbial ecology, physiology, and metabolic potential. This comprehensive perspective is crucial not only for biodiversity assessment and environmental monitoring but also for leveraging these microorganisms in sustainable biotechnological applications, ranging from biofuel production to pharmaceutical compound discovery. As global change accelerates, such integrative research frameworks will become increasingly essential to mitigate the ecological and economic risks posed by altered marine and freshwater ecosystems.
The evolution from morphological descriptions to systems biology has unveiled the hidden complexity and functional diversity of cyanobacteria and microalgae. Emerging technologies, including robotic sampling, pressure-retaining transport, rapid filtration, AFM, Cryo-EM, and MicroED, have provided unprecedented insights into structure, function, and interspecies interactions. Coupled with meta-analytic data on environmental responses, these approaches enable predictive modeling of growth, toxicity, and metabolite production under changing climate conditions. By bridging environmental exploration with laboratory analysis, this integrated framework offers both ecological insight and practical avenues for sustainable exploitation of microbial resources, marking a pivotal advancement in microbial biotechnology and environmental science.